In the rapidly evolving field of single-cell RNA sequencing (scRNA-seq), the quest for efficient and accurate clustering methods has become more crucial than ever. Enter scCDCG: a groundbreaking approach that combines deep structural clustering with deep cut-informed graph embedding. This innovative technique promises to revolutionize the way researchers analyze and interpret scRNA-seq data, setting a new standard for precision and efficiency in the world of single-cell genomics.
Introduction to scCDCG Algorithm
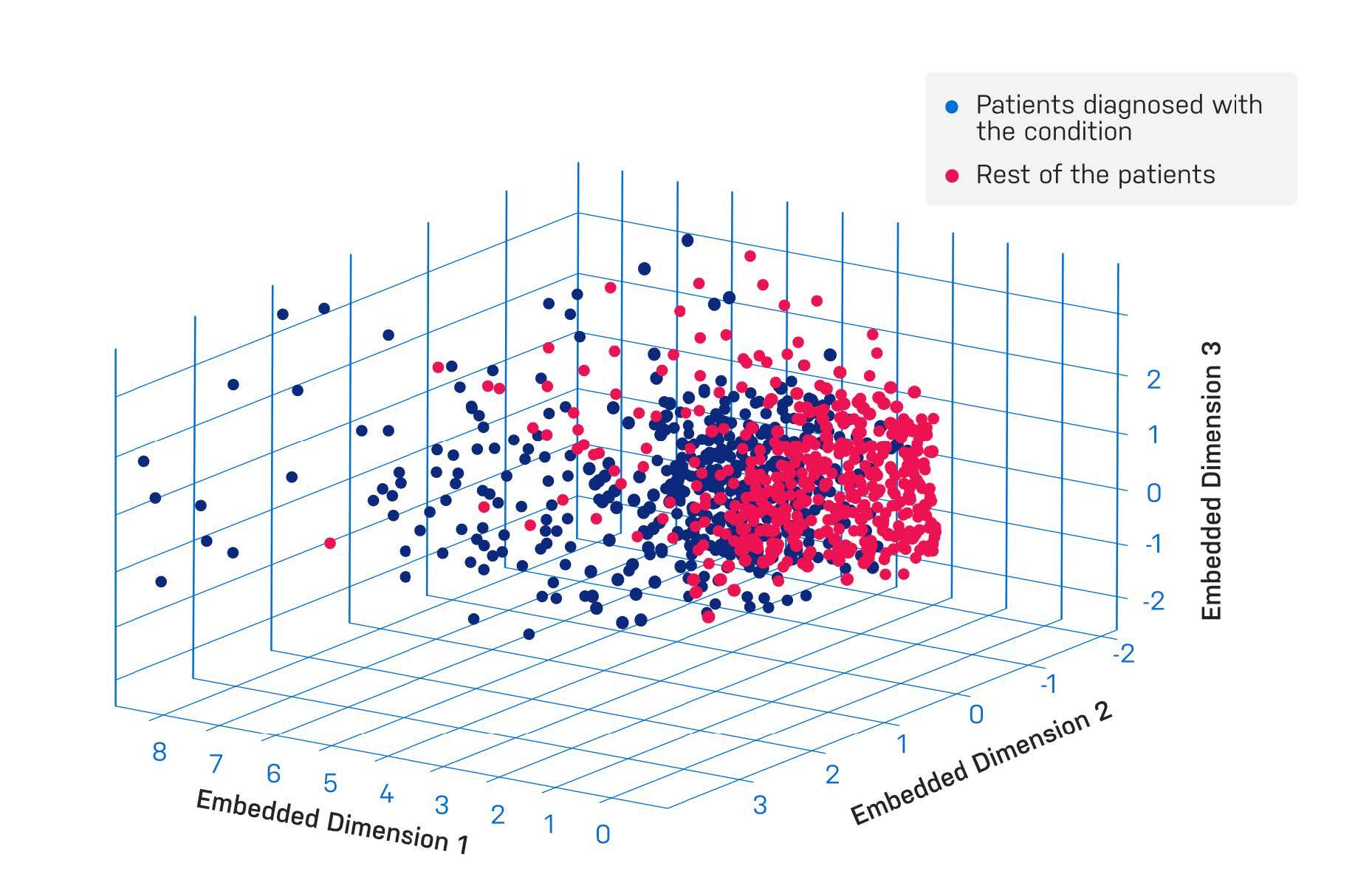
The scCDCG algorithm is a groundbreaking approach to clustering single-cell RNA-seq data efficiently and accurately. By leveraging Deep Cut-informed Graph Embedding, scCDCG can handle the complex and high-dimensional nature of single-cell data, leading to more precise clustering results.
One of the key features of scCDCG is its ability to capture the underlying structure of single-cell RNA-seq data by constructing a graph representation that incorporates deep learning techniques. This allows for the identification of cell populations based on their gene expression profiles, enabling researchers to gain insights into the heterogeneity of cell populations in a more nuanced and informative way. With the scCDCG algorithm, researchers can extract meaningful biological information from single-cell RNA-seq data with greater efficiency and accuracy.
Key Features and Advantages of Deep Cut-informed Graph Embedding
With scCDCG, single-cell RNA-seq data can be efficiently clustered based on deep structural insights provided by our unique Deep Cut-informed Graph Embedding technique. This advanced approach allows for more accurate and robust clustering of cells, uncovering hidden patterns and relationships within the data. By leveraging deep learning and graph embedding, scCDCG offers a powerful tool for researchers to gain deeper insights into the heterogeneity and complexity of single-cell data.
One key feature of scCDCG is its ability to handle large-scale single-cell datasets with high efficiency and accuracy. The Deep Cut-informed Graph Embedding method enables the algorithm to capture intricate relationships between cells, leading to more precise clustering results. Additionally, scCDCG offers advantages such as scalability, interpretability, and versatility, making it a valuable tool for a wide range of single-cell RNA-seq analyses. Researchers can trust scCDCG to provide reliable and informative clustering results, ultimately enhancing their understanding of biological systems at the single-cell level.

Applications and Potential Impact of Efficient Deep Structural Clustering for single-cell RNA-seq
Efficient Deep Structural Clustering for single-cell RNA-seq has the potential to revolutionize the way researchers analyze and interpret complex biological data. With scCDCG, researchers can more accurately identify distinct cell populations within a sample, leading to deeper insights into cellular heterogeneity and gene expression patterns. This innovative approach utilizes Deep Cut-informed Graph Embedding to uncover hidden structures within single-cell RNA-seq data, allowing for more precise clustering and classification of cells.
The applications of scCDCG are vast and impactful, ranging from understanding cellular development and differentiation to identifying biomarkers for diseases. By leveraging the power of deep learning and graph embedding techniques, researchers can efficiently analyze large-scale single-cell RNA-seq datasets, making meaningful discoveries about cell types and their functions. The ability to uncover complex relationships within biological data sets using scCDCG has the potential to accelerate research in fields such as immunology, oncology, and developmental biology.

Future Directions and Recommendations for Further Research
In order to further advance the field of single-cell RNA sequencing and deep learning, future research should focus on the following key areas:
- Exploring the potential of incorporating additional biological features into the scCDCG model to improve clustering accuracy and biological interpretation.
- Investigating the scalability of scCDCG to handle larger single-cell datasets and exploring the possibility of parallel processing to reduce computational time.
- Considering the application of scCDCG to other omics data types beyond single-cell RNA sequencing, such as single-cell ATAC-seq or single-cell proteomics data.
Moreover, it would be valuable to conduct comparative studies with other state-of-the-art algorithms to benchmark the performance of scCDCG and validate its efficacy on various biological datasets. Additionally, exploring the interpretability of the deep cut-informed graph embedding technique and examining the biological relevance of the identified cell clusters would provide further insights into the underlying biological processes captured by the model. As the field of single-cell genomics continues to evolve, the development and refinement of efficient deep learning models such as scCDCG will play a crucial role in advancing our understanding of cellular heterogeneity and dynamics.
Insights and Conclusions
In conclusion, scCDCG offers a groundbreaking approach to deep structural clustering for single-cell RNA-seq data, utilizing deep cut-informed graph embedding to efficiently identify cell subpopulations. By leveraging the power of deep learning and graph theory, this innovative method opens up new possibilities for understanding complex cellular heterogeneity. With its potential to revolutionize the field of single-cell analysis, scCDCG paves the way for exciting advancements in our understanding of cellular biology. Stay tuned as researchers continue to explore and expand upon the capabilities of this cutting-edge technique. Exciting times lie ahead in the world of single-cell research!